6 Machine Learning: Classification
Introduction
In the previous section we covered the concept of distances in euclidean space, and how machines interpret similarity and dissimilarity. We also saw how machines partition datasets into clusters via hierarchical clustering.
In this section, we will cover classification using K-nearest neighbours. Many of the concepts from last week are carried forward to this week, however, the objective of classification tasks is not to perform an exploratory data analysis, but rather we are interested in training a model to correctly place new ‘unseen data’ into the correct group (flower species, cancer type, etc.).
Consider the image below containing a subset of the Iris dataset with a new unknown flower species added to the data:

KNN
K-nearest neighbours operates by identifying the closest neighbouring data points, sorting them by a distance metric and using the top k labelled points to make a decision on the unseen dataset. The choice of k is up to the user i.e how many data points should we consider for voting?
See below the KNN algorithm in operation when k=3:

Operating under the assumption that data within a cluster share similar features, the closest group in euclidean space is the best candidate for the new data. Our new data point thus belongs to the Setosa flower species.
Coding a Classifier
Implementing a KNN model in R is a relatively straightforward call:
knn3(numerical features, feature labels, K)
- numerical features: These are the continuous variables in our dataset.
- feature labels: The labels associated with the samples.
- K: The number of neighbours to consider.
Why do we provide feature labels? Are we essentially giving the algorithm the answers to the test? Yes – but only for the training dataset which is used to train the model. Once the model has been trained, we deploy it on the test dataset to see how well it performs (we can assess the models answers here).
Training & Test Splits
Typically one would use 70% of their data to train the model and 30% to test the model.
We will perform this in R using the Iris dataset:
# 1. Load Iris data, libraries. library(caret) iris <- datasets::iris # 2. Create index for split set.seed(123) train_size <- floor(0.70 * nrow(iris)) train_idx <- sample(seq_len(nrow(iris)), size = train_size) # 3. Dataframe subsetting using index in step 2. train <- iris[train_idx,] test <- iris[-train_idx,]
Training the model
Now that we have created training and test datasets, we will pass the training dataset to the knn3() function.
Recall that the first argument are the numerical variables, followed by the variable labels and a value for K:
# create training model model_fit <- knn3(train[,1:4], train[,5], k = 10)
Now we must deploy the model on the training data (numerical variables) from which it was derived to see how well it performs.
We will use a Confusion Matrix to interpret how well the model did:
model_predictions <- predict(model_fit, train[,1:4], type = "class") train_cfm <- confusionMatrix(model_predictions, train[,5]) print(train_cfm)
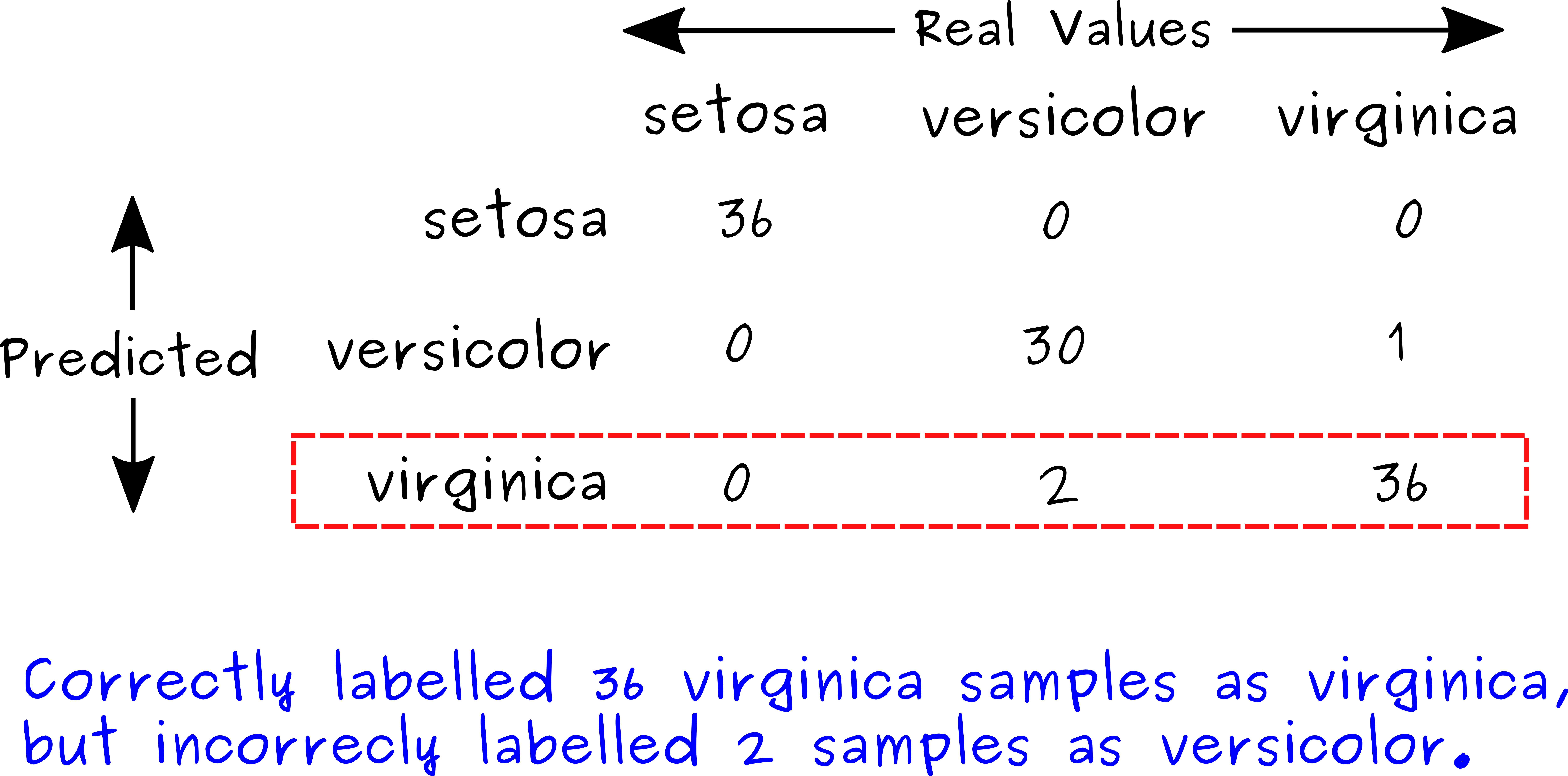
Confusion Matrix and Statistics Reference Prediction setosa versicolor virginica setosa 36 0 0 versicolor 0 30 1 virginica 0 2 36 Overall Statistics Accuracy : 0.9714 95% CI : (0.9188, 0.9941) No Information Rate : 0.3524 P-Value [Acc > NIR] : < 2.2e-16 Kappa : 0.957 Mcnemar's Test P-Value : NA Statistics by Class: Class: setosa Class: versicolor Class: virginica Sensitivity 1.0000 0.9375 0.9730 Specificity 1.0000 0.9863 0.9706 Pos Pred Value 1.0000 0.9677 0.9474 Neg Pred Value 1.0000 0.9730 0.9851 Prevalence 0.3429 0.3048 0.3524 Detection Rate 0.3429 0.2857 0.3429 Detection Prevalence 0.3429 0.2952 0.3619 Balanced Accuracy 1.0000 0.9619 0.9718
Confusion Matrix
Interpretation of the confusion matrix given in the output above:


Testing the model
We have a model that performs very well on the training dataset which is to be expected! Now let’s deploy the model on 30% of the dataset it has never seen before. Like the training model, we will evaluate the accuracy of the model using a confusion matrix.
# Use test dataframe in place of train: test_predictions <- predict(model_fit, test[,1:4], type = "class") # Generate confusion matrix test_cfm <- confusionMatrix(test_predictions, test[,5]) print(test_cfm)
Confusion Matrix and Statistics Reference Prediction setosa versicolor virginica setosa 14 0 0 versicolor 0 17 0 virginica 0 1 13 Overall Statistics Accuracy : 0.9778 95% CI : (0.8823, 0.9994) No Information Rate : 0.4 P-Value [Acc > NIR] : < 2.2e-16 Kappa : 0.9664 Mcnemar's Test P-Value : NA Statistics by Class: Class: setosa Class: versicolor Class: virginica Sensitivity 1.0000 0.9444 1.0000 Specificity 1.0000 1.0000 0.9688 Pos Pred Value 1.0000 1.0000 0.9286 Neg Pred Value 1.0000 0.9643 1.0000 Prevalence 0.3111 0.4000 0.2889 Detection Rate 0.3111 0.3778 0.2889 Detection Prevalence 0.3111 0.3778 0.3111 Balanced Accuracy 1.0000 0.9722 0.9844
Our model is looking pretty good!
Predicting Unseen Data
Now for the real test – let’s use the model on our ‘new flower’ that we saw earlier in this section.
Firstly, we need to create the data point and add it to our test dataset:
# add the new sample new_sample <- data.frame(4.7, 3.5, 2.6, 1, "New Data") names(new_sample) <- colnames(iris) # add it to the test dataset test <- rbind(test, new_sample) # re-deploy model on test dataset: test_predictions <- predict(model_fit, test[,1:4], type = "class") test_cfm <- confusionMatrix(test_predictions, test[,5]) print(test_cfm)
Confusion Matrix and Statistics Reference Prediction setosa versicolor virginica New Data setosa 14 0 0 1 versicolor 0 17 0 0 virginica 0 1 13 0 New Data 0 0 0 0 Overall Statistics Accuracy : 0.9565 95% CI : (0.8516, 0.9947) No Information Rate : 0.3913 P-Value [Acc > NIR] : 4.643e-16 Kappa : 0.9351 Mcnemar's Test P-Value : NA Statistics by Class: Class: setosa Class: versicolor Class: virginica Class: New Data Sensitivity 1.0000 0.9444 1.0000 0.00000 Specificity 0.9688 1.0000 0.9697 1.00000 Pos Pred Value 0.9333 1.0000 0.9286 NaN Neg Pred Value 1.0000 0.9655 1.0000 0.97826 Prevalence 0.3043 0.3913 0.2826 0.02174 Detection Rate 0.3043 0.3696 0.2826 0.00000 Detection Prevalence 0.3261 0.3696 0.3043 0.00000 Balanced Accuracy 0.9844 0.9722 0.9848 0.50000
Inspect the confusion Matrix – under the New Data column, which row has the sample been placed in? (where there is a 1). Does this agree with our guess made about the species of this newly collected flower?