2 RStudio Cloud
Introduction
R is a free software environment for statistical computing and graphics. It compiles and runs on a wide variety of UNIX platforms, Windows and MacOS. Fortunately, there exists a free cloud-based version of R available to both students and teachers at R Studio Cloud. This cloud-based version of RStudio is platform-agnostic – the only requirement being an active internet connection and a web browser.
Follow the steps below to register an account on R Studio Cloud and create a compute environment.
Create Account
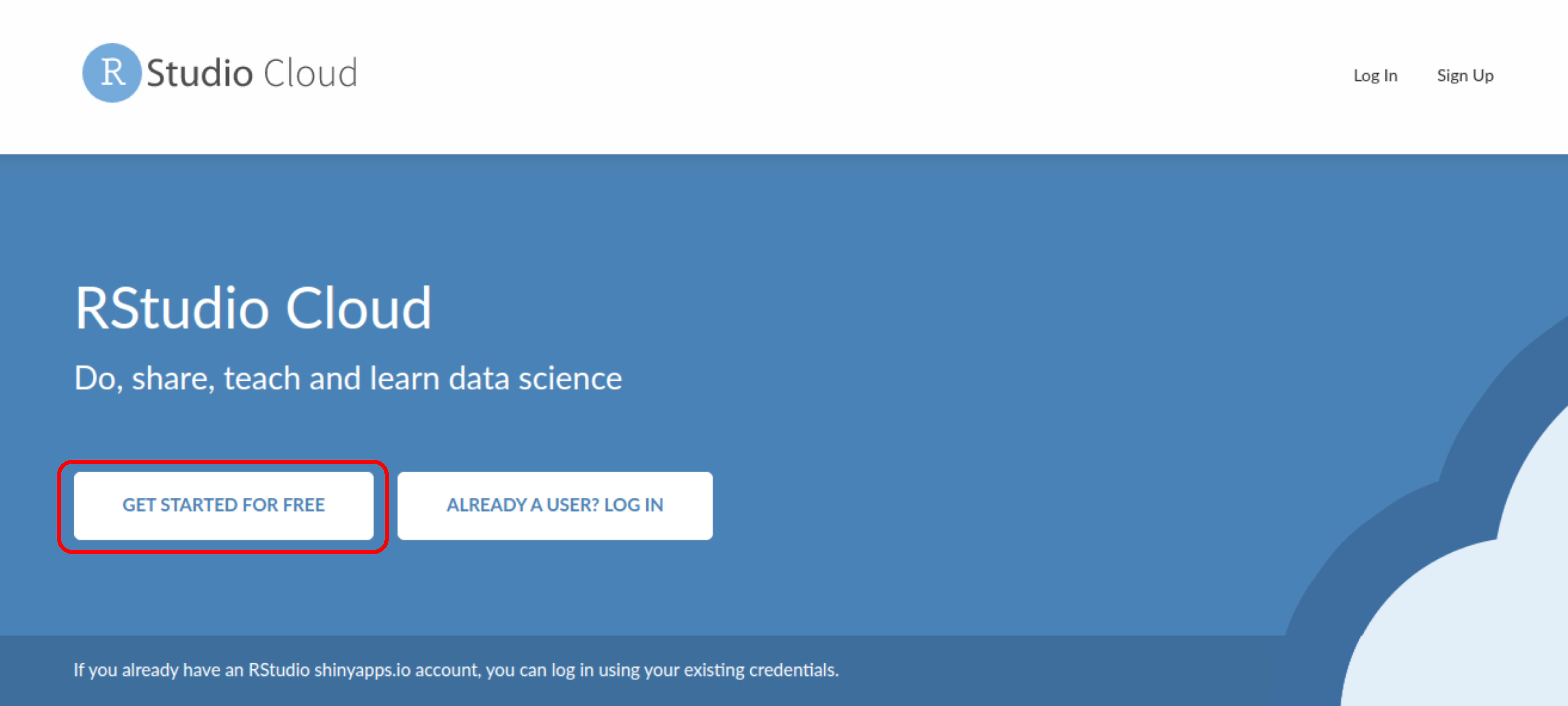
Navigate to the R Studio Cloud website and click on the GET STARTED FOR FREE button.
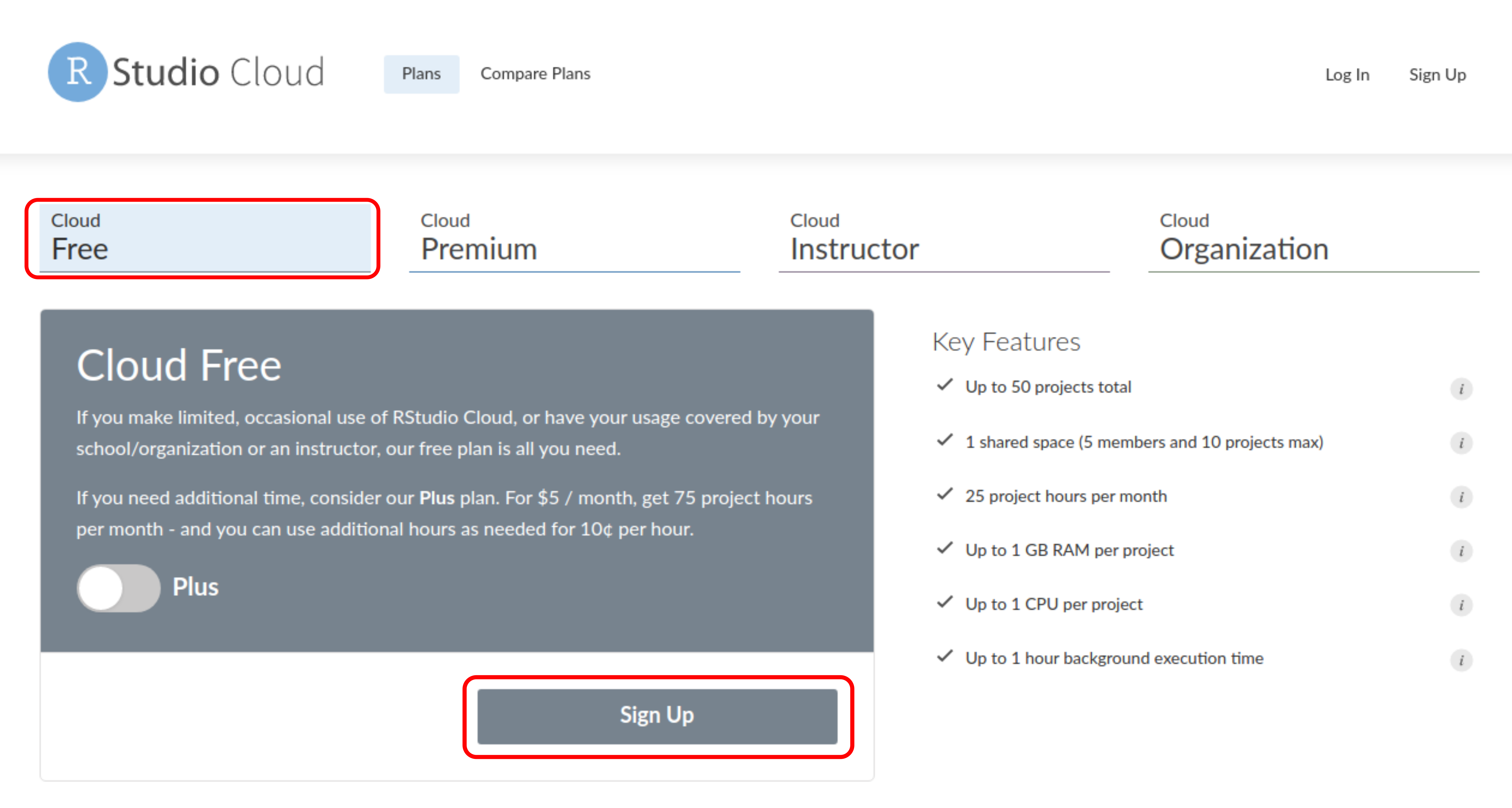
Select a free account. This will provide us with enough resources to perform the operations in this book (1 GB RAM, 1 CPU and 25 hours per month). In reality, this is a very small resource allocation – many computational operations in genomics require exponentially more resources which are performed on high performance compute (HPC) clusters or more recently, on cloud platforms.
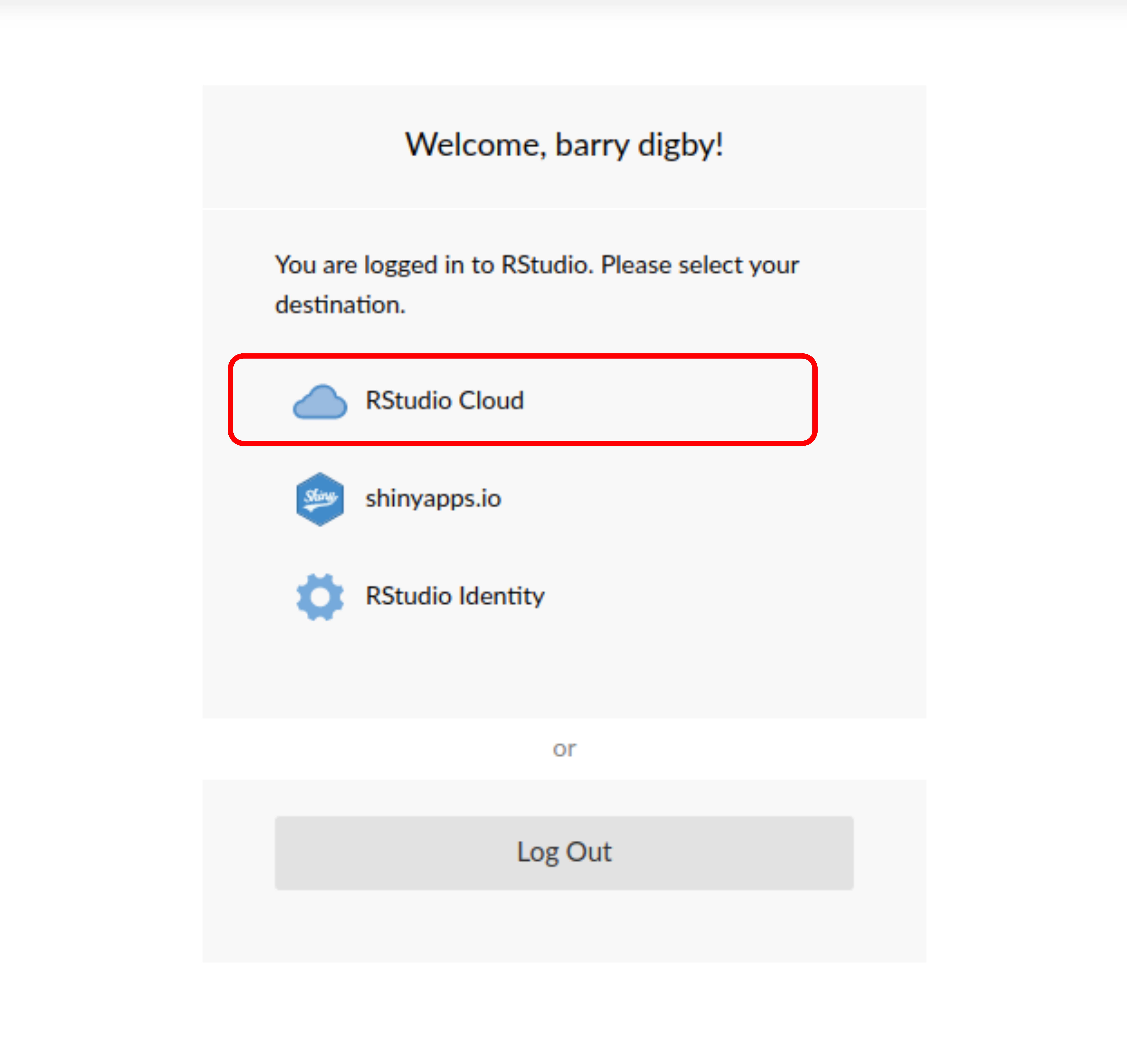
Enter a valid email address and fill out the rest of the information (password, name). RStudio Cloud will send you a verification email with a link to verify your email address. Click on the link in the email to verify your email address – you should now be able to log into RStudio Cloud.
Select RStudio Cloud from the options listed below:
Create Project
Once your account has been created, you need to create a project for the workshop. This is to let RStudio Cloud know what type of environment you want to work in and allows them to track your resource usage.
Select New Project > New R Studio Project:

R Markdown
When analysing datasets in this book we want to be able to create PDF documents containing our code and code outputs. To do this, use the R Markdown document type.
In your project workspace, select File > New File > R Markdown…
You will be prompted to install packages, select yes.

Test R Markdown using a test template:
- Select File > New File > R Markdown…
- Change the Title from ‘Untitled’ to ‘Test’
- Change to Default Output Format to PDF
Your workspace will look like the screenshot below:
- The R Markdown document. We can see the title name ‘Test’ and the preferred output method chosen when setting up the document. The grey lines are known as ‘code-chunks’ which are executable code blocks. To run the code blocks, press the play button. To create a code block, it must be wrapped in (at a minimum) three backticks and the chosen coding language. To finish the code block, insert three further backticks (“`). The contents of the R Markdown document are compiled and converted to a PDF document when the ‘knit’ button is pressed by the user.
- The code console panel. Tracks all lines of previously run code, and can be used to run lines of code if the user chooses to do so.
- The RStudio environment. Keeps track of all objects created by the user. Each item is clickable allowing the user to view the contents of the object.

Install Packages
In your project workspace, you have some very basic packages that come pre-installed in R. We will need to install more packages for the workshop.
Installing packages to your workspace is analogous to installing apps on your phone via the App Store / Google Play except we are installing packages from Bioconductor – a home for open-source bioinformatics software.
In the console (or create a code block), please insert the following lines of code to install the required packages:
install.packages("ggplot2") install.packages("pheatmap") install.packages("ggpubr") install.packages("rcolorbrewer") install.packages("cluster") install.packages("gplots") install.packages("caret") install.packages("e1071")